Abstract
While many patients with follicular lymphoma (FL) have excellent outcomes, early disease progression is associated with poor overall survival (OS). Intensified frontline treatment can improve outcomes in patients considered to be high-risk based on clinical algorithms such as the Follicular Lymphoma International Prognostic Index (FLIPI), however, currently there are no predictive biomarkers to guide frontline risk-adapted therapy. The challenge of predictive biomarker discovery in FL is rooted, in significant part, in its heterogeneity at the genetic level and in cell composition within the tumor microenvironment (TME). To address this, we sought to identify genetic subtypes of FL based on shared transcriptional and microenvironment features.
To characterize the biological heterogeneity of FL at the bulk tumor level, we performed non-negative matrix factorization (NMF) on the microarray transcriptomes of 302 pre-treatment FL cases (GSE127462). All cases were FL grade 1-3a with no additional clinical annotation available and thus the discovery cohort analysis was completely agnostic to clinical outcomes. Four biologically distinct transcriptional subtypes emerged, each characterized by over 1,500 significantly differentially expressed genes (adjusted P-value <0.05) which revealed a spectrum of shared and unique dysregulated biological pathways with unique therapeutic vulnerabilities.
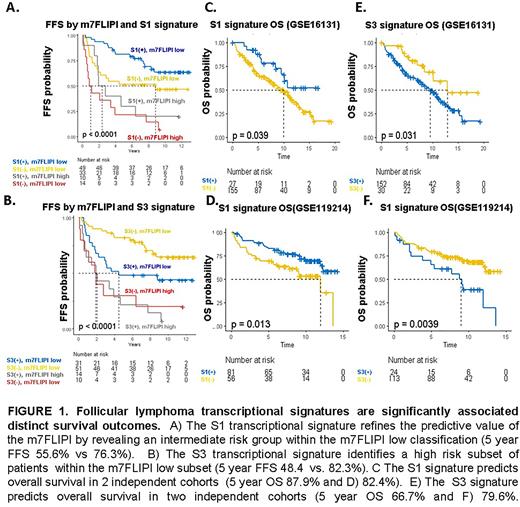
To evaluate the prognostic significance of these 4 signatures, we performed signature recovery on two independent FL datasets annotated with mature clinical data (both with OS, one with failure free survival (FFS), which identified heterogeneous survival outcomes between transcriptional subtypes (Figure 1) and highlighted the potential predictive value of the signatures for treatment naïve patients initiating frontline therapy with rituximab and chemotherapy. In GSE119214 (n=137), all four subtypes were significantly predictive of FFS (FFS HRs and p-values for S1-4 subtypes: 0.59[p=0.03], 0.58[p=0.02], 1.7[0.05], and 2.1[p=0.009], respectively), and three of four were predictive of OS (OS HRs and p-values for S1-4 subtypes: 0.5[p=0.02], 0.6[p=0.06], 2.1[p=0.02], and 2.4[p=0.005]. In a multivariate analysis that included m7FLIPI scores, the S1 signature independently contributed to FFS (p=0.02). These findings were confirmed in an independent dataset (GSE16131; n=182) annotated with OS but not FFS (S1 OS HR 0.5[p=0.04], S3 2.1 [P=0.04]). In a multivariate analysis including age and LDHr, the S01 transcriptional signature independently contributed to OS (HR 0.4[p=0.03]); mutational data was not available in the second dataset, precluding inclusion of m7FLIPI score in multivariate analysis. Signature recovery from an independent microarray dataset with accompanying exome sequencing data (GSE132929, n=65) indicated that the favorable-risk S1 signature was enriched for EZH2 alterations (p=0.02) while the S2 signature was associated with infrequent BCL2 mutations (p=0.002) and the adverse-risk S3 subtype was enriched for alterations frequently present in ABC-DLBCL(CD79b[p=0.002], BTG2[0=0.02], PRDM15[p=0.002]).
To determine the TME features associated with the FL signatures identified in bulk transcriptome analyses and gain insight into the role of specific TME cell types in shaping FL clinical behavior, we performed digital cytometry analyses (Ecotyper) using a novel lymph node signature matrix that included unique lymph node microenvironment stromal populations prevalent in FL including follicular dendritic cells (FDCs), fibroblastic reticular cells (FRCs), lymphatic endothelial cells (LECs), and blood endothelial cells (BECs). De novo transcriptional signature discovery was performed on the discovery dataset (GSE127462) and identified 55 transcriptionally-distinct TME immune and stromal cell states which had unique associations with FL transcriptional subtypes, of which at least 12 were consistently predictive of OS across two separate independent patient cohorts (GSE119214 GSE16131).
In summary, our analyses uncovered genetic subtypes of FL with distinct biological and clinical features, providing a potential framework for advancing precision medicine strategies in FL. These results suggest the possibility of using transcriptional signatures and/or their surrogates as novel biomarkers to guide front-line risk-adapted therapy.
Disclosures
Diefenbach:BMS: Consultancy, Research Funding; Celgene: Consultancy; FATE Therapeutics: Research Funding; Genentech/Roche: Consultancy, Research Funding; Genmab: Consultancy; Gilead: Current equity holder in publicly-traded company; IMAB: Consultancy; Incyte: Research Funding; Kite: Consultancy; MEI Pharma: Research Funding; Merck: Consultancy, Research Funding; MorphoSys: Consultancy, Research Funding; Seattle Genetics: Research Funding.
Author notes
∗Asterisk with author names denotes non-ASH members.